Knowledgebase Integration
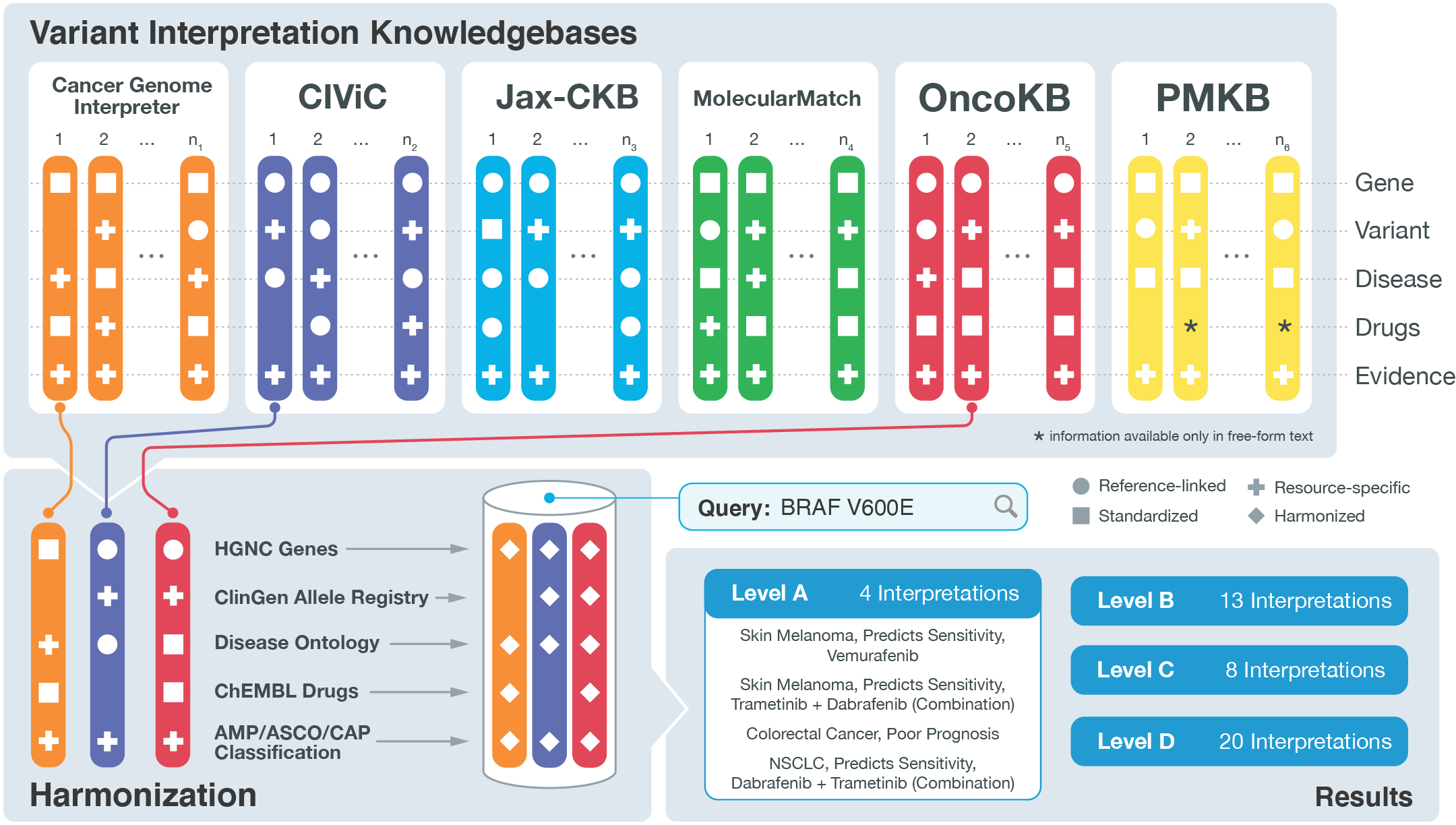
Aggregating Genomic Variant Interpretations
The availability of high-throughput, low-cost sequencing has transformed the landscape of biomedical research by dramatically expanding our capacity to interrogate the sequence of the human genome. Consequently, there has been an explosion of biomedical literature describing the role of specific genomic variants and their impact on human diseases. The six knowledgebases of the VICC have been independently created to curate the biomedical literature for these interpretations. However, the vast majority of the cited papers from any of the knowledgebases are unique to the collective. These findings illustrate the enormity of the task of curating the biomedical literature.
The knowledgebase integration project originated from the GA4GH Genotype-to-Phenotype framework. The intent of the project is to leverage the collective knowledge of the disparate existing resources of the VICC to improve the comprehensiveness of clinical interpretation of genomic variation. An ongoing goal will be to provide and improve upon standards and guidelines by which other groups with clinical interpretation data may make it accessible and visible to the public. Our initial harmonization effort (the VICC Meta-Knowledgebase) and associated analysis was recently described in our flagship manuscript in Nature Genetics.
We continue to actively develop the metakb, and are currently evaluating new sources for inclusion and working on a new search interface. It is currently available online at https://search.cancervariants.org/.
Publications
- A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer - Nature Genetics
- FHIR Genomics: enabling standardization for precision medicine use cases - npj Genomic Medicine
Project Leaders
- Alex H. Wagner, PhDNationwide Children's Hospital, The Ohio State University
- Brian Walsh, BSOregon Health & Science University
- Xuelu "Jeff" Liu, PMP, CSMDana Farber Cancer Institute


